Kulathu Lab
Deubiquitinating enzymes - Regulators of ubiquitin signals:
Deubiquitinases (DUBs) are “erasers” of ubiquitylation that remove ubiquitin from modified proteins to modulate the strength and duration of ubiquitin signals. Given their key roles as regulators of ubiquitylation, there is growing interest to explore DUBs as therapeutic targets. Indeed many DUBs are deregulated in cancers and inflammation. However, the precise biological roles and identity of substrates of many DUBs is unknown. We are therefore interested in exploring the biological roles and how DUBs are targeted to their substrate.
The human genome encodes for around 100 DUBs that are classified into five different DUB families. Many of the DUBs are well studied and it has long been accepted that all DUBs are annotated. We recently made the surprising discovery of DUB activity in FAM63A, an unstudied protein with a domain of unknown function. Based on our analyses, we defined FAM63A to belong to a new family of DUBs that we named MINDY (MIU containing novel DUB family). The catalytic MINDY domain adopts a conformation that is distinct from other known DUBs. In humans, the MINDY family is composed of FAM63A/MINDY-1, FAM63B/MINDY-2, FAM188A/MINDY-3 and FAM188B/MINDY-4.
Interestingly, all MINDY DUBs are highly selective at cleaving K48-linked polyubiquitin chains. MINDY DUBs are highly conserved in evolution and are present in all eukaryotes. However, virtually nothing is known about their biological function.

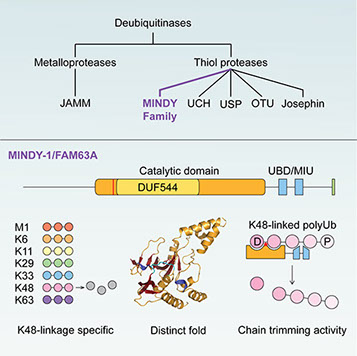
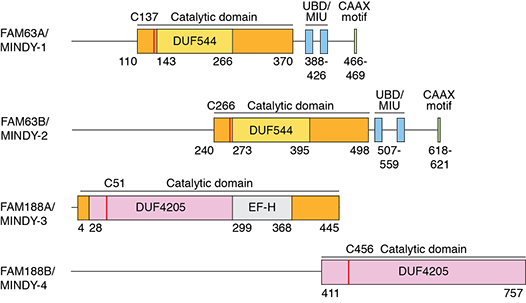
Each of the MINDY DUBs show distinct subcellular localization and we hypothesize localization-specific roles for MINDY DUBs in regulating protein degradation. We use structural approaches to understand how MINDY DUBs achieve substrate recognition. We are using genetic and biochemical approaches to dissect the biological roles of MINDY DUBs and to understand how MINDY DUBs regulate protein degradation.
We discovered ZUFSP/ZUP1 to form the 7th class of DUBs (Paper in Molecular Cell). ZUP1 is selective at cleaving K63 chains and is important for maintaining genome stability. We are studying how ZUP1 achieves substrate specificity.
With growing interest in targeting DUBs in various diseases our mechanistic insights will contribute to these efforts. With thousands of ubiquitylation modifications in a cell and only 100 DUBs to regulate them all, a major question is how DUBs achieve substrate specificity. We aim to identify cellular substrates of DUBs and understand how substrate targeting is achieved.
Back to Research Interests