Northwood
Project with
One of the key challenges in molecular biology is to understand how post-translational modification events regulate protein function. In the Maniaci lab, we aim to uncover how proteases generate new protein fragments and how these fragments contribute to cellular homeostasis and disease development. Proteolysis is an irreversible post-translational modification that not only regulates protein stability but also creates fragments with distinct activities, interactions, and localisation1. Pathogens and cancer cells hijack proteolysis to advance their own agendas by degrading host proteins, disrupting cellular defenses, and promoting invasion and metastasis.
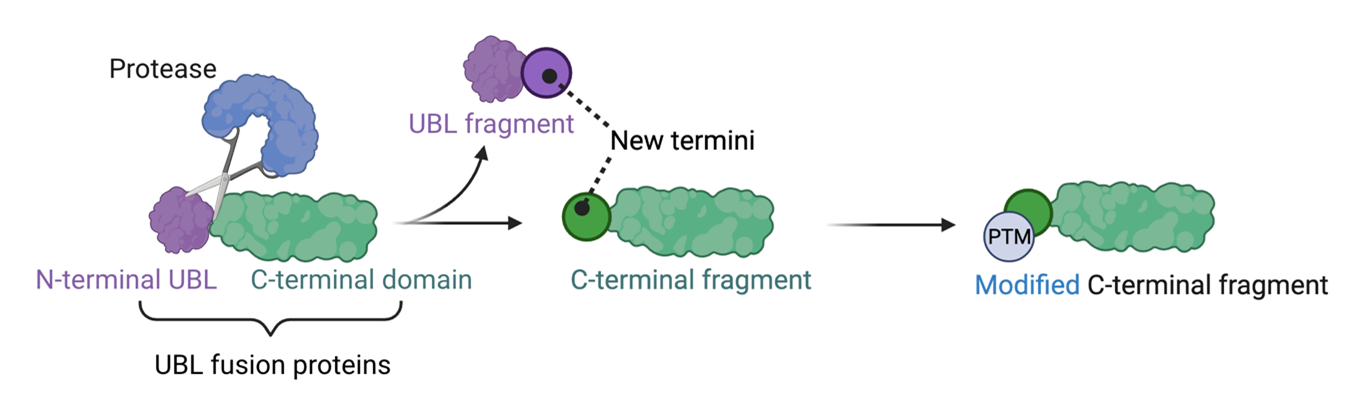
A striking example of this process comes from the ubiquitin-like domain (UBL) fusion proteins2. These proteins undergo proteolytic processing as a means of controlling their physiological roles3. We have recently discovered that the resulting fragments can be further modified with unprecedented post-translational modifications, a mechanism we refer to as “cleave-to-modify” (Figure1). This represents an entirely new layer of proteome regulation. However, very little is known about the UBL fusion protein family, the proteases responsible for their cleavage, or the downstream biological consequences of this mechanism.
This PhD project aims to identify novel substrates and pathways regulated by the “cleave-to-modify” mechanism and link these discoveries to cellular function. By doing so, the project will provide fundamental insights into how proteolytic processing diversifies protein function and reveal new opportunities for therapeutic intervention.
This project takes advantage of our recently developed toolkit to study UBL fusion proteins and their processing4. Students interested in gaining expertise in a wide variety of approaches are strongly encouraged to apply, since the project merges several disciplines, including: method development, state-of-the-art mass spectrometry, cell biology, biophysics, and biochemistry.
The Maniaci lab is a young, dynamic, and collaborative research group with a strong focus on mentoring and professional development. As a PhD student, you will receive hands-on training from your supervisor and senior lab members, benefit from regular lab meetings, journal clubs, and one-to-one guidance, present your work at national and international conferences, interact with a wide network of collaborators across academia and industry, and develop transferable skills in critical thinking, communication, and project management.
Our lab is committed to fostering an inclusive and supportive environment where creativity and curiosity are encouraged. This studentship offers the chance to make exciting discoveries at the forefront of protein biology while building a strong foundation for a successful research career.

References
- Lange, P. F. & Overall, C. M. in Current Opinion in Chemical Biology Vol. 17 73-82 (Elsevier Ltd, 2013).
- Kerscher, O., Felberbaum, R. & Hochstrasser, M. in Annu. Rev. Cell Dev. Biol Vol. 22 159-180 (2006).
- Thakran, P. et al. in The EMBO Journal Vol. 37 89-101 (John Wiley & Sons, Ltd, 2018).
- Hales, L.T., Tammiste P.M., […] Maniaci C. A Proteolytic Switch: USP5 controls SDE2 function via UBL-directed cleavage. doi: https://doi.org/10.1101/2025.05.23.655772

