Our cells are continually subjected to damaging stress from both external sources, like UV radiation, and internal sources, such as free radicals generated by malfunctioning mitochondria. Fortunately, the body has several mechanisms to manage this stress. One crucial protective process is autophagy—an intracellular, membrane-based lysosomal degradation pathway. Autophagy safeguards cellular health by removing defective or toxic components that could otherwise accumulate and cause harm. Due to this vital role, impairment of autophagy can have serious consequences and is associated with numerous diseases, including cancer, neurodegenerative disorders, and cardiovascular disease.
Our research aims to uncover how to control this still not fully understood pathway, with the goal of leveraging autophagy as a therapeutic strategy. This requires identifying the physiological signals that induce autophagy and understanding how these signals lead to autophagosome formation. Our work is particularly centred on the ULK1 protein kinase complex, which is essential for initiating autophagy, though its exact mechanism remains unclear. The ULK1 complex comprises several subunits and promotes autophagy through both kinase-dependent and kinase-independent mechanisms. It is thought to act as a key signaling hub that integrates various upstream cues—such as nutrient and energy status sensed via mTOR and AMPK pathways—into the initiation of autophagosome biogenesis. Clarifying how these signals are coordinated and the cellular responses they elicit is a major focus of our lab.
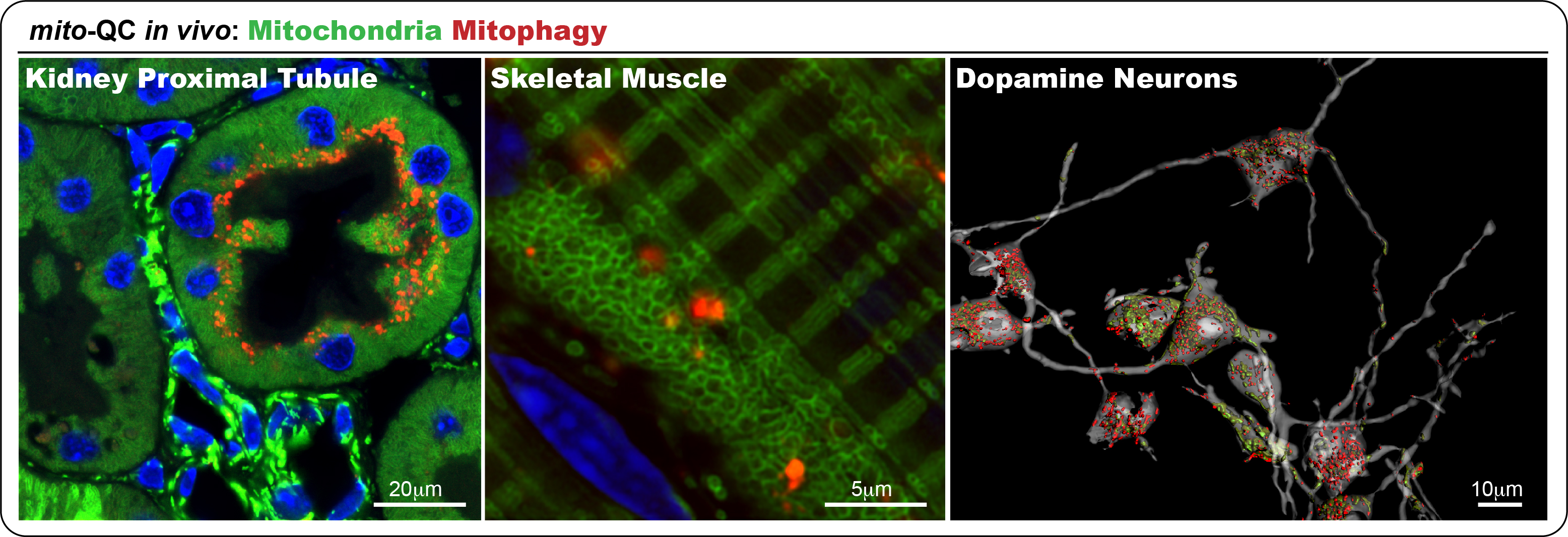
Recently, we developed the mito-QC reporter model to monitor mitophagy, the selective autophagic removal of mitochondria, which is disrupted in conditions like Parkinson’s disease. Through these models, we have identified multiple physiological instances of mitophagy, including within midbrain dopaminergic neurons. Our ongoing objectives are to characterize the underlying mechanisms, map the involved signalling pathways, and explore how these processes relate to development and disease progression. Notably, our recent studies show that the most common Parkinson’s-associated mutation, LRRK2 G2019S, impairs mitophagy in vivo. Importantly, this defect can be corrected using small molecule LRRK2 inhibitors, offering encouraging prospects for future therapeutic development.
Diversity is key in all aspects of our work, ranging from the type of experiments we do, to the members of the lab. This enriches our science, productivity and inclusiveness. Whatever your background, we try to foster an open-minded approach to honestly answer important scientific questions about autophagy. Interested in joining us? Informal enquires can be made by e-mailing Ian Ganley (i.ganley@dundee.ac.uk).



