The end of chromosome replication
One of the most striking features of chromosome replication in eukaryotic cells is that it only happens once in each cell cycle. This makes DNA replication very different to transcription, during which each gene is copied many times into RNA. Whereas an RNA polymerase can melt double-strand DNA on its own, a DNA polymerase must wait for its template to be provided by the action of a DNA helicase, which unwinds the DNA duplex and represents a key focus for regulation. The replicative helicase in eukaryotic cells is highly complicated and is controlled in an exquisite fashion, so that it only gets one single opportunity to unwind each stretch of the genome during every round of chromosome replication. In a new paper just published in Science, Marija Maric from Karim Labib's group has identified an exciting new mechanism by which the eukaryotic replicative helicase is regulated.
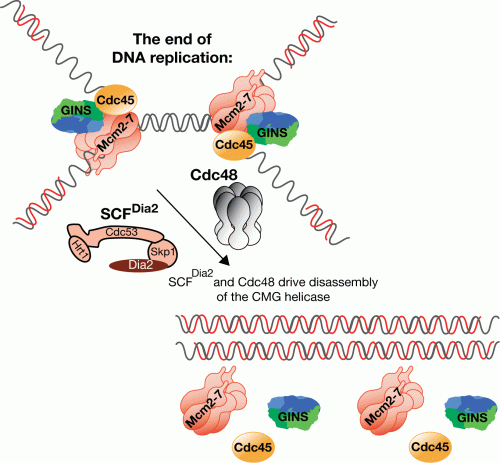
The essential helicase at eukaryotic forks is a multimeric complex known as ‘CMG' or Cdc45-MCM-GINS. The catalytic core of the helicase is formed by the six Mcm2-7 proteins, which are loaded around DNA in an inactive form during the G1-phase of the cell cycle. Once cells enter S-phase, the loaded Mcm2-7 hexamers are activated by recruitment of two missing components known as Cdc45 and ‘GINS' (itself a complex of four small proteins). The activation process is driven by cyclin dependent kinase and Cdc7 kinase, which phosphorylate the Mcm2-7 helicase core, as well as two of the three loading factors that recruit GINS and Cdc45 to origins of replication. Work over the last decade has shown that the helicase activation mechanism is conserved from humans to yeast, indicating that it represents a universal mechanism for the initiation of chromosome replication in eukaryotic cells.
Once activated, the CMG helicase is very tightly associated with DNA replication forks and must never be displaced, as reloading or reactivation cannot occur until the subsequent cell cycle. Nevertheless, when two converging replication forks from neighbouring origins meet each other, replication is terminated in that region of the genome, and disassembly of the CMG helicase is likely to be the key regulated step in replisome dissolution. Until now, nothing was known about the underlying mechanism of helicase disassembly.
Following up on an original observation from a former postdoc in Karim's group, Giacomo de Piccoli, Marija showed that the CMG helicase is ubiquitylated, specifically on its Mcm7 subunit, in a reaction that only occurs at the end of chromosome replication. Ubiquitylation of the helicase leads to a disassembly reaction that requires the Cdc48/p97 segregase, which probably acts by pulling the ubiquitylated Mcm7 subunit out of the CMG helicase, causing the rest of the complex to fall apart. The E3 ligase responsible for this process is called SCFDia2 and was identified by Timurs Maculins during his PhD studies in Karim's group. Tim showed that SCFDia2 is essential for disassembly of the CMG helicase at the end of S-phase, consistent with Marija's data. These findings raise many interesting questions that will drive new projects in the replication field over the coming years. In addition to elucidating fully the mechanism of helicase disassembly, a key issue will be to address how the process is regulated so that it never occurs before the termination of replication, but then always happens. By analogy with the initiation of chromosome replication, it will be important to explore whether helicase disassembly also involves a universal mechanism in diverse eukaryotic species, which helps to preserve genome integrity in proliferating cells. Whatever the answer, it now seems clear that the end of chromosome replication is regulated just as carefully as the start.
To read a copy of Marija's paper that has just been published, click here.
The essential helicase at eukaryotic forks is a multimeric complex known as ‘CMG' or Cdc45-MCM-GINS. The catalytic core of the helicase is formed by the six Mcm2-7 proteins, which are loaded around DNA in an inactive form during the G1-phase of the cell cycle. Once cells enter S-phase, the loaded Mcm2-7 hexamers are activated by recruitment of two missing components known as Cdc45 and ‘GINS' (itself a complex of four small proteins). The activation process is driven by cyclin dependent kinase and Cdc7 kinase, which phosphorylate the Mcm2-7 helicase core, as well as two of the three loading factors that recruit GINS and Cdc45 to origins of replication. Work over the last decade has shown that the helicase activation mechanism is conserved from humans to yeast, indicating that it represents a universal mechanism for the initiation of chromosome replication in eukaryotic cells.
Once activated, the CMG helicase is very tightly associated with DNA replication forks and must never be displaced, as reloading or reactivation cannot occur until the subsequent cell cycle. Nevertheless, when two converging replication forks from neighbouring origins meet each other, replication is terminated in that region of the genome, and disassembly of the CMG helicase is likely to be the key regulated step in replisome dissolution. Until now, nothing was known about the underlying mechanism of helicase disassembly.
Following up on an original observation from a former postdoc in Karim's group, Giacomo de Piccoli, Marija showed that the CMG helicase is ubiquitylated, specifically on its Mcm7 subunit, in a reaction that only occurs at the end of chromosome replication. Ubiquitylation of the helicase leads to a disassembly reaction that requires the Cdc48/p97 segregase, which probably acts by pulling the ubiquitylated Mcm7 subunit out of the CMG helicase, causing the rest of the complex to fall apart. The E3 ligase responsible for this process is called SCFDia2 and was identified by Timurs Maculins during his PhD studies in Karim's group. Tim showed that SCFDia2 is essential for disassembly of the CMG helicase at the end of S-phase, consistent with Marija's data. These findings raise many interesting questions that will drive new projects in the replication field over the coming years. In addition to elucidating fully the mechanism of helicase disassembly, a key issue will be to address how the process is regulated so that it never occurs before the termination of replication, but then always happens. By analogy with the initiation of chromosome replication, it will be important to explore whether helicase disassembly also involves a universal mechanism in diverse eukaryotic species, which helps to preserve genome integrity in proliferating cells. Whatever the answer, it now seems clear that the end of chromosome replication is regulated just as carefully as the start.
To read a copy of Marija's paper that has just been published, click here.

