Researchers from Adrien Rousseau’s lab in the MRC Protein Phosphorylation and Ubiquitylation Unit at the University of Dundee, Scotland, have identified mechanisms contributing to the stress-induced increase in proteasome abundance using yeast. This new study is now published in Nature Cell Biology.
When cells are stressed, they initiate stress responses in order to survive. These responses vary with the stress, but typically involve widespread turnover of the proteome. Proteins which are either damaged or no longer required are degraded, while proteins used by the cell to fight against the stress are rapidly produced. Increasing protein degradation increases the pool of free amino acids, used to produce stress response proteins, and removes potentially harmful damaged proteins. Cells increase protein degradation, in part, by increasing proteasome abundance. Cancer cells, which are subject to multiple stresses, often have high levels of proteasomes, while age-related diseases frequently see reduced proteasome levels and a corresponding increase in damaged proteins.
Prior to starting his research group at the MRC PPU, Adrien Rousseau identified a protein kinase, Mpk1, which, when activated, acts to increase proteasome assembly. Mpk1 does this by increasing the translation of proteasome assembly chaperones, which then in turn help construct more proteasomes. Exactly how this increase in translation occurred remained mysterious. The Rousseau Lab has recently identified that actin remodelling controls the translation of these proteasome assembly chaperones.
Adrien and a visiting masters student, Roberta Cacioppo, hypothesised that proteins bound to proteasome assembly chaperone mRNA would help increase their translation following stress. To test this hypothesis, they performed a mass spectrometry screen in yeast with Houjiang Zhou to identify proteins bound to translating proteasome assembly chaperone mRNA. One of the proteins they identified, endocytic protein Ede1, does exactly that. This unexpected discovery required further mechanistic dissection.
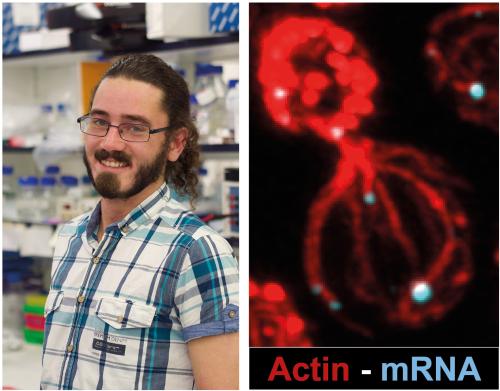
To determine how Ede1 contributes to mRNA translation, postdoctoral research associate Tom Williams performed further screening and characterisation using high-resolution microscopy, genetic screening, and mRNA localisation and cytoskeletal manipulations. Using these techniques, additional proteins which are important for Ede1’s interaction with the mRNA were found and the team showed that Ede1 recruits mRNA to highly branched actin structures, termed cortical actin patches. This recruitment happens when the actin cytoskeleton is disrupted, a frequent early response to stress. Recruitment to cortical actin patches is critical for increased mRNA translation. Tom was able to bypass the requirement for Ede1 by tethering proteasome assembly chaperone mRNA directly to the cortical actin patch, revealing Ede1’s role in their translation.
The team is now investigating further mechanisms of proteasome assembly chaperone mRNA localisation and translation initiation signals, and retains an interest in identifying further mRNAs that may be translated at highly branched actin structures.
To read a copy of the manuscript describing these findings click here

