Nic Dzamko and Jeremy Nichols report findings that will help understanding the function of LRRK2 and development of inhibitors
Autosomal dominant missense mutations within the gene encoding for the Leucine-Rich Repeat protein Kinase 2 (LRRK2) predispose humans to develop Parkinson's disease. The most frequent mutation (G2019S) enhances kinase activity suggesting inhibitors may be useful for the treatment of Parkinson's disease.
Numerous drug companies have developed drugs that inhibit LRRK2, but as no downstream substrates of this enzyme have been identified there has been no way to evaluate the effectiveness of these compounds to suppress LRRK2 activity in cell or animal systems.
Nic Dzamko and Jeremy Nichols working in the Alessi lab have published two papers in the Biochemical Journal that provide clues as to how LRRK2 functions and will be of use to drug companies developing inhibitors of LRRK2 for the treatment of Parkinson's disease. Nic and Jeremy first discovered that LRRK2 is phosphorylated at two residues (Ser910 and Ser935) enabling it to bind isoforms of 14-3-3 [1]. They observed that disrupting 14-3-3 binding by mutating Ser910 and Ser935 caused LRRK2 to accumulate within cytoplasmic aggregates resembling inclusion bodies, rather than being diffusely localised throughout the cytoplasm [1]. Interestingly, Jeremy also observed that five of the six most common pathogenic LRRK2 mutations (namely R1441C, R1441G, R1441H, Y1699C, I2020T) displayed markedly reduced phosphorylation of Ser910/Ser935 and 14-3-3 binding and hence were localised within inclusion bodies [1]. These results suggest that certain pathogenic mutations in LRRK2 disrupt ability of LRRK2 to be phosphorylated at Ser910 and Ser935 through an as yet unknown mechanism that we will investigate in future work.
Another key striking observation was that treatment of cells with two structurally diverse LRRK2 inhibitors (H1152 and sunitinib) induced rapid dephosphorylation of Ser910 and Ser935, resulting in loss of 14-3-3 binding and accumulation of LRRK2 within cytoplasmic inclusion bodies [2]. Nic has found in unpublished work that all other much more specific LRRK2 inhibitors that we have tested also induce the dramatic dephosphorylation of LRRK2 at Ser910/Ser935 leading to loss of 14-3-3 binding and causing LRRK2 to localise within inclusion bodies. Furthermore a mutant of LRRK2 that is resistant to H1152 and Sunitinib termed LRRK2[A2016T] does not become dephosphorylated or localise to inclusion bodies in cells treated with LRRK2 inhibitors [2].
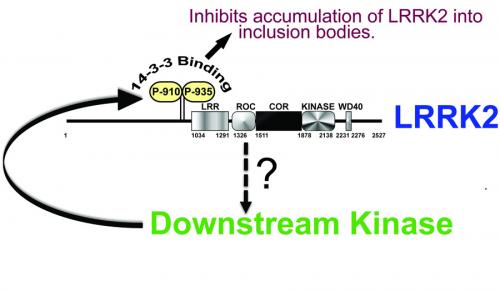
Nic and Jeremy's data strongly suggests that LRRK2 does not autophosphorylate at Ser910 or Ser935. The key idea that we are exploring is whether LRRK2 operates as an upstream kinase to activate an as yet unknown downstream kinase that phosphorylates Ser910/Ser935 as part of a negative feedback regulatory loop. Our model of how Ser910/Ser935 phosphorylation is regulated is summarised in the figure.
Our recent results should be of great interest to pharmaceutical companies as they indicate that monitoring phosphorylation of LRRK2 at Ser910 and Ser935 or 14-3-3 binding as well as studying diffuse versus aggregated LRRK2 cytoplasmic localisation can be used to evaluate the relative potency of LRRK2 inhibitors. These assays could be deployed in cell lines, tissues of animals or humans treated with LRRK2 inhibitors. Moreover, for human patients administered LRRK2 inhibitors, phosphorylation status of LRRK2 at Ser910 and Ser935 in blood cells could perhaps be employed as a biomarker of LRRK2 inhibitor activity.
The results described in these papers will also hopefully stimulate future work aimed at understanding how Ser910 and Ser935 phosphorylation is regulated. It would be fascinating to explore whether the Ser910/Ser935 protein kinase might also comprise a drug target for the treatment of Parkinson's disease.
1 Nichols, R. J., Dzamko, N., Morrice, N. A., Campbell, D. G., Deak, M., Ordureau, A., Macartney, T., Prescott, A. R. and Alessi, D. R. (2010) 14-3-3 binding regulates LRRK2 cytoplasmic localisation and is disrupted by multiple Parkinson's disease associated mutations. In press in the Biochemical Journal (click here to read paper)
2 Dzamko, N., Deak, M., Henati, F., Reith, A. D., Prescott, A. R., Alessi, D. R. and Nichols, R. J. (2010) Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser910/Ser935, disruption of 14-3-3 binding and altered cytoplasmic localisation. In press in the Biochemical Journal (click here to read paper)
Numerous drug companies have developed drugs that inhibit LRRK2, but as no downstream substrates of this enzyme have been identified there has been no way to evaluate the effectiveness of these compounds to suppress LRRK2 activity in cell or animal systems.
Nic Dzamko and Jeremy Nichols working in the Alessi lab have published two papers in the Biochemical Journal that provide clues as to how LRRK2 functions and will be of use to drug companies developing inhibitors of LRRK2 for the treatment of Parkinson's disease. Nic and Jeremy first discovered that LRRK2 is phosphorylated at two residues (Ser910 and Ser935) enabling it to bind isoforms of 14-3-3 [1]. They observed that disrupting 14-3-3 binding by mutating Ser910 and Ser935 caused LRRK2 to accumulate within cytoplasmic aggregates resembling inclusion bodies, rather than being diffusely localised throughout the cytoplasm [1]. Interestingly, Jeremy also observed that five of the six most common pathogenic LRRK2 mutations (namely R1441C, R1441G, R1441H, Y1699C, I2020T) displayed markedly reduced phosphorylation of Ser910/Ser935 and 14-3-3 binding and hence were localised within inclusion bodies [1]. These results suggest that certain pathogenic mutations in LRRK2 disrupt ability of LRRK2 to be phosphorylated at Ser910 and Ser935 through an as yet unknown mechanism that we will investigate in future work.
Another key striking observation was that treatment of cells with two structurally diverse LRRK2 inhibitors (H1152 and sunitinib) induced rapid dephosphorylation of Ser910 and Ser935, resulting in loss of 14-3-3 binding and accumulation of LRRK2 within cytoplasmic inclusion bodies [2]. Nic has found in unpublished work that all other much more specific LRRK2 inhibitors that we have tested also induce the dramatic dephosphorylation of LRRK2 at Ser910/Ser935 leading to loss of 14-3-3 binding and causing LRRK2 to localise within inclusion bodies. Furthermore a mutant of LRRK2 that is resistant to H1152 and Sunitinib termed LRRK2[A2016T] does not become dephosphorylated or localise to inclusion bodies in cells treated with LRRK2 inhibitors [2].
Nic and Jeremy's data strongly suggests that LRRK2 does not autophosphorylate at Ser910 or Ser935. The key idea that we are exploring is whether LRRK2 operates as an upstream kinase to activate an as yet unknown downstream kinase that phosphorylates Ser910/Ser935 as part of a negative feedback regulatory loop. Our model of how Ser910/Ser935 phosphorylation is regulated is summarised in the figure.
Our recent results should be of great interest to pharmaceutical companies as they indicate that monitoring phosphorylation of LRRK2 at Ser910 and Ser935 or 14-3-3 binding as well as studying diffuse versus aggregated LRRK2 cytoplasmic localisation can be used to evaluate the relative potency of LRRK2 inhibitors. These assays could be deployed in cell lines, tissues of animals or humans treated with LRRK2 inhibitors. Moreover, for human patients administered LRRK2 inhibitors, phosphorylation status of LRRK2 at Ser910 and Ser935 in blood cells could perhaps be employed as a biomarker of LRRK2 inhibitor activity.
The results described in these papers will also hopefully stimulate future work aimed at understanding how Ser910 and Ser935 phosphorylation is regulated. It would be fascinating to explore whether the Ser910/Ser935 protein kinase might also comprise a drug target for the treatment of Parkinson's disease.
1 Nichols, R. J., Dzamko, N., Morrice, N. A., Campbell, D. G., Deak, M., Ordureau, A., Macartney, T., Prescott, A. R. and Alessi, D. R. (2010) 14-3-3 binding regulates LRRK2 cytoplasmic localisation and is disrupted by multiple Parkinson's disease associated mutations. In press in the Biochemical Journal (click here to read paper)
2 Dzamko, N., Deak, M., Henati, F., Reith, A. D., Prescott, A. R., Alessi, D. R. and Nichols, R. J. (2010) Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser910/Ser935, disruption of 14-3-3 binding and altered cytoplasmic localisation. In press in the Biochemical Journal (click here to read paper)

