My research aims to understand how quantitative phosphoinositide 3-kinase (PI3K) signalling underpins context-dependent information transmission in mammalian cells, and to use this knowledge to identify novel therapeutic options for diseases of aberrant PI3K signalling. I am committed to achieving this aim through:
- Research with integrity (https://ukcori.org/what-research-integrity-is/)
- Transparent sharing of all our results without unnecessary delays (https://ralitsamadsen.wordpress.com/open-science/)
- Care and respect for others (https://ralitsamadsen.wordpress.com/teaching/)
At present, my research is driven by an unmet need for better therapies for PIK3CA-related disorders. It is my conviction that this need can only be met through a transition from a largely qualitative to a fully quantitative understanding of PI3K signalling.
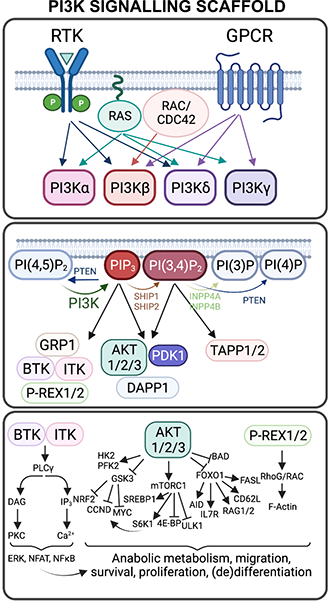
 The human PIK3CA gene codes for the catalytic subunit of the ubiquitously expressed phosphoinositide 3-kinase alpha (PI3Kα), an enzymatic complex that is crucial for transducing signals from growth factor-specific receptor tyrosine kinases, including IGF1R, INSR, EGFR and many others. Recent work by myself and others has demonstrated that the landscape and biochemical consequences of genetic PI3Ka activation are more complex than previously appreciated. For example, a heterozygous, activating PIK3CA-H1047R mutation results in increased PI3K signalling but no overt phenotypic reprogramming of human induced pluripotent stem cells; in contrast, homozygous expression of the same variant in the same cellular context results in transcriptomic rewiring, constitutive stemness and pluripotency loss. Evidence for dose-dependent effects of PI3K signalling activation, with relevance for disease outcome, can also be found in publicly available breast cancer datasets. Collectively, these findings hint at the existence of poorly defined biochemical thresholds which, along with short-and long-term temporal control of signalling activity, determine the phenotypic output of aberrant PI3K activation in human disease.
The human PIK3CA gene codes for the catalytic subunit of the ubiquitously expressed phosphoinositide 3-kinase alpha (PI3Kα), an enzymatic complex that is crucial for transducing signals from growth factor-specific receptor tyrosine kinases, including IGF1R, INSR, EGFR and many others. Recent work by myself and others has demonstrated that the landscape and biochemical consequences of genetic PI3Ka activation are more complex than previously appreciated. For example, a heterozygous, activating PIK3CA-H1047R mutation results in increased PI3K signalling but no overt phenotypic reprogramming of human induced pluripotent stem cells; in contrast, homozygous expression of the same variant in the same cellular context results in transcriptomic rewiring, constitutive stemness and pluripotency loss. Evidence for dose-dependent effects of PI3K signalling activation, with relevance for disease outcome, can also be found in publicly available breast cancer datasets. Collectively, these findings hint at the existence of poorly defined biochemical thresholds which, along with short-and long-term temporal control of signalling activity, determine the phenotypic output of aberrant PI3K activation in human disease.
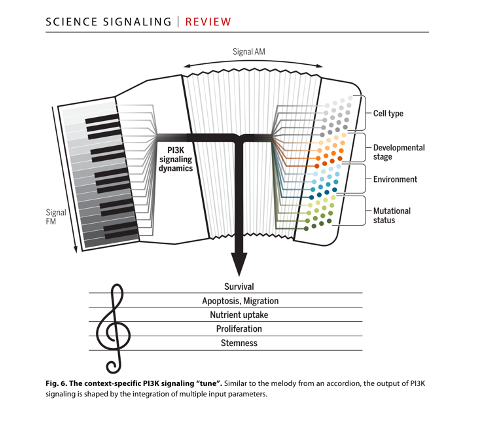
 While the typically static, pictorial representations of signalling pathways in textbooks and reviews may give the impression of linear information transfer and signalling control, the reality is far more complicated. A quantitative understanding of PI3K signalling therefore requires a multidisciplinary approach, featuring a firm grasp of quantitative biochemistry, cell biology, and computational modelling. These facets must be considered in the context of the wider disease biology that we seek to modify, including attention to unintended consequences on normal human physiology. I therefore work according to a classical systems biology approach that covers the following objectives:
While the typically static, pictorial representations of signalling pathways in textbooks and reviews may give the impression of linear information transfer and signalling control, the reality is far more complicated. A quantitative understanding of PI3K signalling therefore requires a multidisciplinary approach, featuring a firm grasp of quantitative biochemistry, cell biology, and computational modelling. These facets must be considered in the context of the wider disease biology that we seek to modify, including attention to unintended consequences on normal human physiology. I therefore work according to a classical systems biology approach that covers the following objectives:
- The development of quantitative signalling maps (akin to electronic circuits) that depict how the PI3K network transduces a specific environmental signal in different cell types. Using quantitative set-ups (2D/3D single cell signalling, live-cell microscopy of PI3K activity) developed during my Sir Henry Wellcome Fellowship, alongside computational and mathematical biology, our experiments seek to decipher the mechanisms used by cells to encode growth factor identity through spatiotemporal PI3K signalling.
The engineering of human induced pluripotent stem cell (iPSC)-derived disease models with controlled expression of activating PIK3CA mutations. Aided by quantitative phenotyping, mathematical modelling and the unique capacity of iPSCs to differentiate into derivatives of all three germ layers (mesoderm, endoderm, ectoderm), these models will be used to: a. understand the molecular mechanisms underpinning the ability of PI3Kα to control cell fate decisions in a dose-dependent manner, including identification of “points-of-no-return” that are no longer reversible by simple PI3Kα inhibition; b. to test candidate therapies identified in (1) in lineage-specific and more disease-relevant settings, with a focus on fine-tuning rather than complete ablation of critical signalling components.