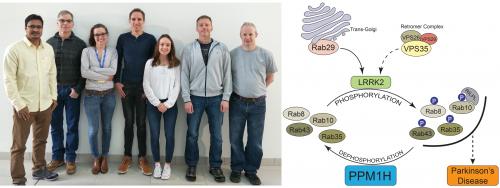
MRC PPU Researchers have been working to better understand how mutations that increase LRRK2 kinase activity cause Parkinson’s disease. LRRK2 phosphorylates a subset of Rab GTPases including Rab8A and Rab10 within a region of the “Switch-II motif” that controls the interaction with effectors such as RILPL1 and RILPL2.
The protein phosphatase(s) that dephosphorylates Rab proteins thereby reversing the impact of LRRK2 is unknown. Our work suggested that this phosphatase(s) may be highly active, as treatment of cell lines with LRRK2 inhibitors induces a rapid dephosphorylation of Rab10.
Francesca Tonelli (Postdoc in Alessi lab) first explored whether the well-studied PP1 and PP2A protein phosphatases play a role in regulating Rab protein phosphorylation, but found they didn’t. This prompted Kerryn Berndsen (PhD student) and Pawel Lis (Postdoc) working in the Alessi lab to undertake a focused siRNA screen of all human protein phosphatases. This uncovered a poorly studied serine/threonine protein phosphatase termed PPM1H, that when knocked down promoted Rab10 phosphorylation.
Kerryn and Pawel, with help from Thomas Macartney (MRC PPU CRISPR Scientist) and Raja Sekhar Nirujogi (Postdoc in Alessi lab), next generated PPM1H knockout cell lines and demonstrated by immunoblot and mass spectrometry analysis that lack of PPM1H increased endogenous Rab8A and Rab10 phosphorylation. Kerryn also found that PPM1H knock-out suppressed Rab10 dephosphorylation following addition of an LRRK2 inhibitor termed MLi-2.
Consistent with PPM1H comprising a Rab phosphatase, Kerryn also found that overexpression of wild-type PPM1H in cells markedly suppressed LRRK2-mediated phosphorylation of Rab3, Rab8, Rab10, Rab35 and Rab43, whereas overexpression of the catalytically inactive PPM1H mutant had no effect on Rab phosphorylation.
A key experiment was then to test whether PPM1H could directly dephosphorylate LRRK2-phosphorylated Rab proteins in vitro. However, to generate sufficient stoichiometrically phosphorylated Rab proteins for these experiments, initially proved very challenging as it was not possible to purify sufficient recombinant LRRK2. This problem was solved by a discovery made by Katie Mulholland (PhD student in Miratul Muqit lab) who found that Rab8A and Rab10 could be efficiently phosphorylated at the same residue that LRRK2 targets, by the MST3 kinase, that is much easier to produce recombinantly than LRRK2 (see here).
Axel Knebel (MRC PPU Protein Scientist) succeeded in elaborating methodology to produce milligram amounts of stoichiometrically Thr72 phosphorylated Rab8A and ~60% Thr73 phosphorylated Rab10. Mark Dorward (MRC Services and Reagent technical Scientist) also elaborated methods to produce wild type and catalytically inactive recombinant PPM1H. These reagents enabled Kerryn to demonstate that PPM1H efficiently and directly dephosphorylated Rab8A and Rab10 in biochemical studies.
Pawel and Kerryn were next able to identify a “substrate-trapping” PPM1H mutant (Asp288Ala) that binds with high affinity to endogenous, LRRK2-phosphorylated Rab8A and Rab10 proteins, under conditions in which wild type and other catalytically inactive mutants do not interact. Raja Nirujogi undertook mass spectrometry analysis to establish that LRRK2 phosphorylated Rab8A and Rab10 were main cellular proteins that specifically co-immunoprecipitated with the substrate trapping PPM1H[D288A] mutant.
Wondwossen Yeshaw and Paulina Wawro (postdocs in Suzanne Pfeffer’s lab at Stanford University) observed that PPM1H is mainly localized to the Golgi apparatus. The Pfeffer lab had previously revealed that the LRRK2 pathway blocks primary cilia formation through a pathway involving LRRK2 phosphorylated Rab8 and Rab10 interacting with RILPL1 (see here). Therefore, Wondwossen Yeshaw set out to uncover how siRNA knockdown of PPM1H impacted cilia formation. He found that PPM1H knock-down suppresses primary cilia formation, similar to pathogenic LRRK2.
Taken together these findings suggest that PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins. Enhancers of PPM1H activity or expression could offer a future therapeutic approach to prevent or treat Parkinson’s disease.
We thank the Michael J Fox Foundation for Parkinson’s Research (MJFF) and the UK Medical Research Council for generously supporting this research. To read a copy of the paper describing these findings click here.

