Congratulations to Asad Malik (Phd Student) and Athanasios Karapetsas (ex MRC PPU postdoc, now working in AstraZeneca) for their tremendous work that has elucidated the mechanism of activation of the Leucine-rich-repeat-kinase 1 (LRRK1) protein kinase.
LRRK1 is a close homologue of the LRRK2 protein kinase that is implicated in Parkinson’s disease. Both LRRK1 and LRRK2 are multidomain kinases possessing a ROC-CORA-CORB containing GTPase domain and phosphorylate distinct Rab proteins.
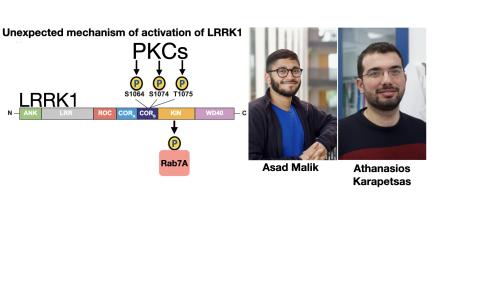
LRRK1 phosphorylates Rab7A at Ser72 within its effector binding motif. Loss of function mutations in LRRK1 cause osteosclerotic metaphyseal dysplasia, a severe bone disorder in humans.
Previous work undertaken by Asad suggested that LRRK1 but not LRRK2, is activated following stimulation of cells with phorbol ester that stimulates the Protein Kinase C (PKC) signalling pathway. How this triggered LRRK1 activation was unknown.
Asad discovered that activation of LRRK1 following phorbol ester stimulation was blocked by several structurally diverse PKC inhibitors including a compound termed Darovasertib that is in human clinical trials for metastatic uveal melanoma. Asad and Athanasios also found that phorbol ester induced phosphorylation of LRRK1 in cells in a manner that was blocked with PKC inhibitors suggesting that PKCs may directly phosphorylate LRRK1.
Asad and Athanasios then showed that in vitro, multiple PKC isoforms were capable of directly phosphorylating and activating recombinant LRRK1 generated by Deep Chatterjee, Sebastian Mathea and Stefan Knapp at the University of Frankfurt.
We assumed that PKCs would activate LRRK1 by phosphorylating the kinase domain which is the common mechanism by which protein kinases are activated. Instead, peptide mapping studies undertaken with the help of Raja Nirujogi and Nick Morrice using a new type of mass spectrometer that is optimised for phospho-peptide analysis (ZenoTOF 7600) revealed that PKCα phosphorylates a cluster of conserved residues (Ser1064, Ser1074 and Thr1075) located within a region of the GTPase domain termed the CORB domain.
Thr1075 lies within an optimal PKC site phosphorylation motif and its mutation to Ala, completely blocked PKC-mediated activation of LRRK1. A triple Glu mutation of Ser1064/Ser1074/Thr1075 to mimic phosphorylation also enhanced LRRK1 kinase activity ~3-fold.
From analysis of available structures of LRRK1 and LRRK2 we speculate that phosphorylation of Ser1064, Ser1074 and Thr1075 could activate LRRK1 by promoting interaction and stabilization of the aC-helix on the kinase domain that may lie close by.
Our work reveals new insights into how the activity of LRRK1 is controlled by PKC phosphorylation and reveals a unique activation mechanism involving phosphorylation of the GTPase domain.
In future work structural analysis will need to be undertaken to define how phosphorylation of LRRK1 impacts kinase activity and whether this is mediated by regulation of GTP binding and/or GTPase activity. It would also be interesting to investigate whether other AGC kinases that are related to PKCs and possess similar substrate specificity would also be capable of activating LRRK1. The role that LRRK1 mediated Rab7A phosphorylation plays also needs to be elucidates
It also seems that this mechanism of activation is unique to LRRK1, as LRRK2 is not activated by PKC phosphorylation and the LRRK1 Ser1064, Ser1074 and Thr1075 phosphorylation sites are not conserved in LRRK2. Data so far indicates that LRRK2 is regulated by Rab proteins binding to its N-terminal ARM domain that is not conserved in LRRK1.
We also thank our colleagues Toan Phung, Melanie Wightman and Robert Gourlay who also helped with this study.
This study was supported the joint efforts of The Michael J. Fox Foundation for Parkinson’s Research (MJFF) and Aligning Science Across Parkinson’s (ASAP) initiative. MJFF administers the grant (ASAP-000463-Alessi lab and ASAP-000159-Knap lab) on behalf of ASAP and itself. The D.R.A lab is also supported by the UK Medical Research Council [grant number MC_UU_00018/1] and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit.
To read a copy of the paper click here

