“Cleave-to-Modify”: Illuminating the “dark proteome” with novel chemical biology tools
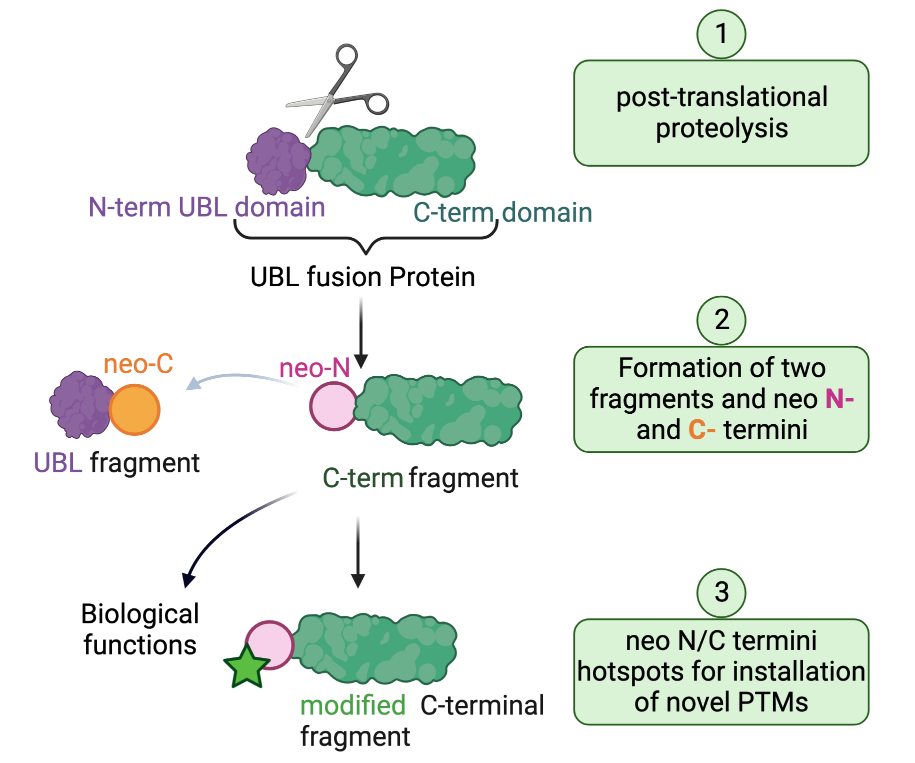
Variations in protein structure at the level of amino acid sequence and modifications allow for the biological complexity observed in living organisms; a complexity that far exceeds that of the approximately 20,000 genes in the human genome that encode for proteins. Post-translational modifications (PTMs) add to the diversity of the proteome through the generation of new and different proteoforms. Amongst the known PTMs, proteolytic cleavage (or proteolysis) is an irreversible modification performed by a family of specialized enzymes termed proteases. It is characterized by the highly precise cleavage of a protein into two or more smaller fragments defined by distinct new ends generated by the protease at the cleavage site, called neo-N and neo-C-termini. These new fragments can be stable in vivo and can exhibit altered biological activity, location, and interactions compared with their precursor protein. However, the mechanisms by which these processes are regulated remain largely mysterious.
An example of proteolytically processed proteins are the members of the ubiquitin-like domain fusion protein family. These undergo proteolytic cleavage as a mean to regulate their stability and physiological role. We have recently discovered that the generated neo-termini serve as “hot spots” for unprecedented PTMs, a mechanism we refer to as “cleave to modify”. However, not much is known about ubiquitin-like domain fusion proteins, the protease enzymes involved in their processing and the biological events and modifications that are actioned as a result. The long-term objective of the Maniaci Lab is to develop tailor-made tools to illuminate and understand the events that lead to the proteolysis of the “dark proteome” of ubiquitin-like domain fusion proteins and their modifications.
Current research focus:
(1) Unravel the key players involved in the “cleave to modify” mechanism
(2) Identify and characterise the PTMs involved and their functional consequences
(3) Illuminate the biology that is actioned as a result of these events
Often regarded as a mere mechanism for protein destruction, proteolytic cleavage has instead a much more widespread regulatory role in fundamental cellular processes like cellular growth, differentiation, apoptosis, and survival. Consequently, misregulation of proteolytic activities is implicated in many severe acute and chronic human diseases. For these reasons, proteases are considered as attractive drug targets while their cleavage products may provide specific disease biomarkers. Therefore, understanding the proteolytic mechanism and the identity, role and repertoire of the substrates and proteolytic products is not only of fundamental interest but might also reveal invaluable information on the neo-biology of reactivities and recognition, underpinning an unexplored layer of complexity in the hierarchy of cell regulation. This knowledge could therefore be used to identify novel disease markers and allow for the development of specialized therapeutics that target only specific proteoforms.
Values
We are committed to establish a supporting environment where lab members can think freely, express themselves openly, and exchange ideas with mutual respect to enable creative thinking and innovation. This commitment will promote an equal, inclusive, and diverse community to drive scientific progress. We welcome applicants from underrepresented and disadvantaged backgrounds. Please get in touch if you are interested in joining the team.



