Motivated by the lack of a molecular understanding and a treatment that slows down the progression of Parkinson’s disease, our research focuses on the leucine-rich repeat kinase-2 (LRRK2) pathway, as mutations that increase the kinase activity of LRRK2, represent one of the most common inherited causes of Parkinson’s disease. The goal of our work is to undertake high quality rigorous research to develop a robust framework of knowledge of how the LRRK2 signalling pathways is regulated, to develop improved methods to better diagnose and treat Parkinson’s disease.
LRRK2 is a large multidomain protein consisting of two catalytic domains namely an atypical GTPase and a protein kinase domain. Pathogenic mutations that activate LRRK2 are found in the kinase and GTPase as well as other domains and are thought to destabilise the inactive conformation of LRRK2 1. We have found that LRRK2 phosphorylates a subgroup of Rab GTPase proteins (Rab1, Rab3, Rab8, Rab10, Rab12, Rab29, Rab35, and Rab43) 2,3, that coordinate membrane homeostasis and endocytic and exocytic pathways. LRRK2 phosphorylates Rab substrates at a conserved Ser or Thr site lying at the centre of the effector-binding switch-II motif (Thr72 for Rab8A, Thr73 for Rab10, and Ser106 for human Rab12) 2,3. We also discovered that this reaction is counteracted by the PPM1H, a highly selective protein phosphatase that efficiently dephosphorylates Rab proteins 4,5.
A summary of our current understanding of the LRRK2 signalling pathway based on our research and that of others is presented in Figure 1.
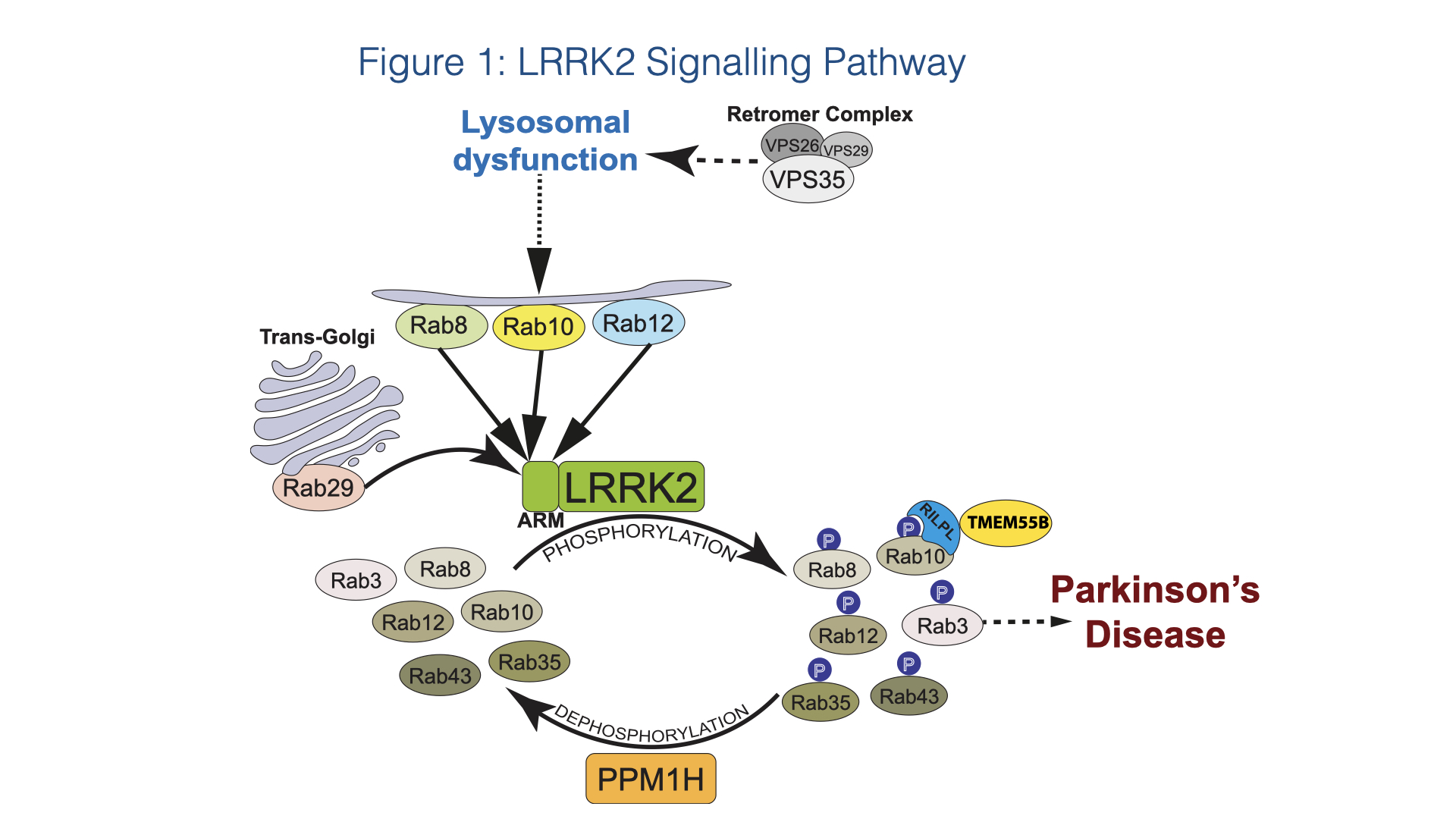
We have identified 4 scaffolding proteins, namely RILPL1, RILPL2, JIP3 and JIP4, that specifically interact with LRRK2 phosphorylated Rab8A and Rab10 with higher affinity than dephosphorylated Rab proteins 2,6. The interaction of RILPL1 with phosphorylated Rab proteins interferes with ciliogenesis 2 and in cholinergic neurons in the striatum leads to disruption of a Sonic hedgehog neuro-protective circuit that supports dopaminergic neurons, providing a pathway by which LRRK2 may be linked to Parkinson’s disease pathology 7.
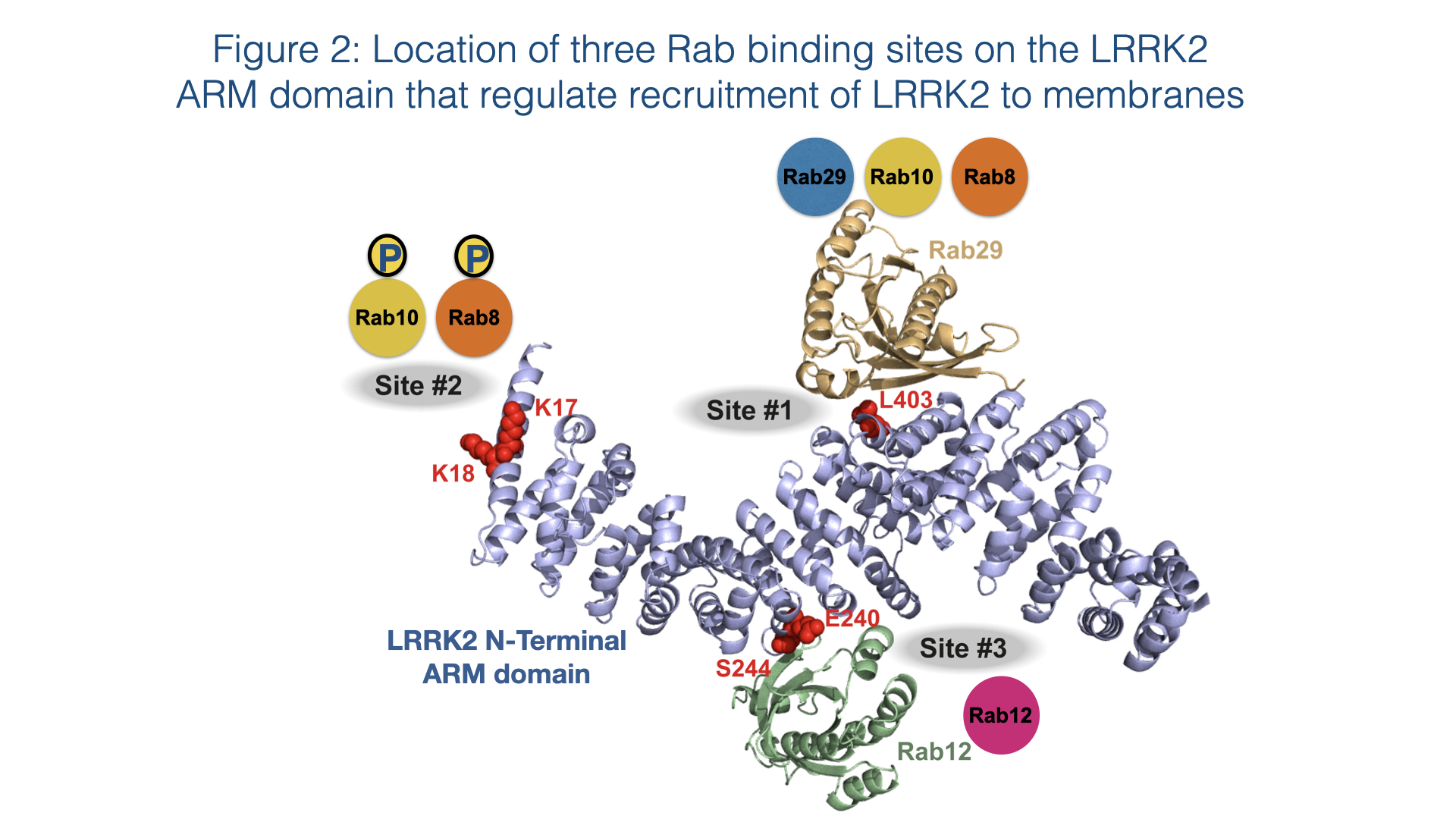
Rab proteins are not only substrates but also play a key role in controlling LRRK2 kinase activity by binding to the N-terminal ARM domain and recruiting LRRK2 to membranes where it becomes activated 8,9. Recent work has pinpointed 3 Rab binding sites within the LRRK2 ARM domain (Figure 2). Site-1 binds to dephosphorylated Rab29, Rab32 as well as Rab8 and possibly Rab10 10,11. Site-2 interacts specifically to LRRK2-phosphorylated Rab8 and Rab10 in a feed-forward pathway that drives membrane recruitment and activation of LRRK2 10. Site-3 interacts with Rab12 and ablation of this site or knock-out of Rab12 has the largest effect in regulating the basal activity of LRRK2 12.
The D620N mutation in the VPS35 component of the retromer complex transports select endosomal cargo proteins between endosomal compartments and the Golgi and has been linked to Parkinson’s disease and stimulates the LRRK2 pathway via an unknown mechanism 13. VPS35[D620N] knock-in cells and tissues display markedly enhanced Rab phosphorylation to a higher level than observed with LRRK2 pathogenic mutations 13,14 and this mutation has been proposed to lead to lysosomal dysfunction. Our on-going work suggests that the D620N VPS35 mutation disrupts selective endosomal cargo trafficking triggering lysosomal dysfunction, thereby activating LRRK2 (Figure 1).
Key questions for on-going work
We wish to define the mechanism by which LRRK2 is recruited to the stressed/damaged lysosome and the role that LRRK2 plays once recruited to the lysosome. We have identified a novel pathway involving LRRK2 controlling the binding of the phospho Rab protein to RILPL1 and the lysosomal protein TMEM55B, that wish to study further. We are also keen to uncover other proteins that interact with LRRK2 phosphorylated Rab proteins including Rab12 and characterise these further. We are also keen to undertake unbiased screens to identify new regulators of the LRRK2 signalling pathway and use this knowledge to develop improved biomarkers and therapeutic strategies to better diagnose and treat LRRK2 driven Parkinson’s disease.
It is exciting times in this research field as late phase clinical trials of LRRK2 inhibitors are underway and targeting this pathway is one of the most promising therapeutic strategies to slow progression of Parkinson’s disease.
Collaboration, open science, culture and mentorship
Alessi lab has a strong track record of fostering collaborations and conducting open science. This includes his Parkinson’s disease research with the Michael J Fox Foundation for Parkinson’s Research and Aligning Science Across Parkinson’s as well as the UK Dementia Research Initiative. He contributes to general research on protein phosphorylation and ubiquitylation by directing the MRC Protein Phosphorylation and Ubiquitylation Unit and supports pharmaceutical companies with their research in this field through the Division of Signal Transduction Unit and other collaborations.
To help the global research community Alessi lab promotes sustainable science practices by freely sharing all reagents and services via the https://mrcppureagents.dundee.ac.uk/ and other reagents websites such as LRRK2.bio. All of Alessi’s lab protocols are now deposited on Protocols.io and papers submitted to preprint servers such as Biorxiv at or before the time of submission. He is also participating on a new UK "National Asset" program to enable all MRC Unit- and Institute-generated tools, data, and technologies to be made available from a web database.
The Alessi lab aims to undertake high quality research, promote a strong sharing culture between researchers and help disseminate tools/data/technologies to better enable scientists to replicate data and support worldwide research.
We are strong supporters of working hard to continuously improve equality and diversity in a multidisciplinary environment, paying attention to cultivating culture and development best practices.
Dario has trained over 30 graduate students and 40 postdoctoral researchers, the majority now in leadership positions in academia and industry.
Louis-Jeantet Foundation video showcasing Alessi lab and its research
Video - Dario Alessi, winner of the 2023 Jeantet-Collen Prize for Translational Medicine
Dario celebrated as winner of the Pritzker Prize for Leadership in Parkinson’s Research
Video - Honoring the Winner of The Michael J. Fox Foundation's 2023 Major Research Prize
Transcript from Michael J Fox Podcast Interview by Marie McNeely
PDF - Interview transcript from Michael J Fox Podcast
For a full updated publication list of Alessi lab publications please visit Google Scholar:
Dario Alessi on Google Scholar
Dario Alessi - Declaration of interests
References
- Kalogeropulou, A. F. et al. Impact of 100 LRRK2 variants linked to Parkinson's disease on kinase activity and microtubule binding. Biochem J 479, 1759-1783 (2022). https://doi.org/10.1042/BCJ20220161
- Steger, M. et al. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. Elife 6, e31012 (2017). https://doi.org/10.7554/eLife.31012
- Steger, M. et al. Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 5, e12813 (2016). https://doi.org/10.7554/eLife.12813
- Berndsen, K. et al. PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins. eLife 8, e50416 (2019). https://doi.org/10.7554/eLife.50416
- Waschbüsch, D., Berndsen, K., Knebel, A., Alessi, D. R. & Khan, A. R. Structural basis for the specificity of PPM1H phosphatase for Rab GTPases. bioRxiv, 2021.2002.2017.431620 (2021). https://doi.org/10.1101/2021.02.17.431620
- Waschbüsch, D. et al. Structural Basis for Rab8a Recruitment of RILPL2 via LRRK2 Phosphorylation of Switch 2. Structure 28, 406-417.e406 (2020). https://doi.org/10.1016/j.str.2020.01.005
- Dhekne, H. S. et al. A pathway for Parkinson's Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. Elife 7, e40202 (2018). https://doi.org/10.7554/eLife.40202
- Purlyte, E. et al. Rab29 activation of the Parkinson's disease-associated LRRK2 kinase. EMBO J 37, 1-18 (2018). https://doi.org/10.15252/embj.201798099
- Liu, Z. et al. LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum Mol Genet 27, 385-395 (2018). https://doi.org/10.1093/hmg/ddx410
- Vides, E. G. et al. A feed-forward pathway drives LRRK2 kinase membrane recruitment and activation. Elife 11 (2022). https://doi.org/10.7554/eLife.79771
- Zhu, H., Tonelli, F., Alessi, D. R. & Sun, J. Structural basis of human LRRK2 membrane recruitment and activation. bioRxiv, 2022.2004.2026.489605 (2022). https://doi.org/10.1101/2022.04.26.489605
- Dhekne, H. S. et al. Genome-wide screen reveals Rab12 GTPase as a critical activator of pathogenic LRRK2 kinase. bioRxiv, 2023.2002.2017.529028 (2023). https://doi.org/10.1101/2023.02.17.529028
- Mir, R. et al. The Parkinson's disease VPS35[D620N] mutation enhances LRRK2-mediated Rab protein phosphorylation in mouse and human. The Biochemical journal 475, 1861-1883 (2018). https://doi.org/10.1042/BCJ20180248
- Nirujogi, R. S. et al. Development of a multiplexed targeted mass spectrometry assay for LRRK2-phosphorylated Rabs and Ser910/Ser935 biomarker sites. Biochem J 478, 299-326 (2021). https://doi.org/10.1042/BCJ20200930



