Parkinson’s disease is a progressive disorder of movement and coordination due to the specific loss of dopaminergic neurons in the brain. There are no medicines that can slow or halt the disease process largely due to lack of understanding of the fundamental mechanisms. Spectacular advances in genetics have identified 13 genes that are mutated in familial forms of the disease and nearly 90 gene variants that increase susceptibility to sporadic disease. Understanding the regulation and signaling networks in which these genes reside is likely to elaborate new knowledge on the key cellular pathways that are vital for maintaining neuronal integrity and health [1]. It may also lead to the development of novel approaches to better diagnose and treat Parkinson’s.
My lab studies the protein kinase, PTEN-induced kinase 1 (PINK1) in which loss of function mutations cause Parkinson’s disease. We are interested in understanding how PINK1 is regulated and how mutations leads to neurodegeneration. Studies to date indicate that PINK1 is a master regulator of a mitochondrial damage response pathway that exists in many different cell types including neurons. Under healthy conditions PINK1 is inactive however, upon selective mitochondrial stress, PINK1 is activated and stimulates the removal of damaged mitochondrial via autophagy (termed mitophagy) thereby promoting cell survival.
My lab found that PINK1 phosphorylates ubiquitin and Parkin at a Ser residue (Serine 65) located within a conserved fold of ubiquitin and the ubiquitin-like domain of Parkin. We also determined the mechanism of activation of Parkin E3 ligase activity by phosphorylated-ubiquitin [2]. We were able to demonstrate the relevance of our findings to the pathogenesis of Parkinson’s through analysis of a Parkin Ser65Ala knockin mouse and the discovery of rare Parkinson’s patients harbouring mutations of the Parkin Ser65 site [3]. In collaboration with the van Aalten lab we solved the first crystal structure of PINK1 using an insect orthologue that has revealed the mechanisms of how disease mutations impact on PINK1 activity [4]. Collectively our work has stimulated interest amongst industry to develop small molecule activators of Parkin and PINK1.
We have also found that PINK1 stimulates the phosphorylation of a subset of Rab GTPases (Rab1B, 8A, 8B and 13) at a conserved Ser residue (Ser111) that lies within the SF3 motif [5]. PINK1 dependent phosphorylation of Rabs disrupts its ability to interact with its cognate guanine exchange factor, Rabin8, and more recently we have found that phosphorylation impairs the ability of another Parkinson’s associated kinase LRRK2 to phosphorylate Thr72 within the Switch II domain [6].
Outstanding questions include understanding how PINK1 is activated upon mitochondrial depolarisation and determining novel regulatory proteins required for PINK1 activation. We are establishing robust methods to study PINK1 signalling in neurons and defining the downstream biology in these cell types. We are also investigating the mechanisms by which Rab GTPases are regulated by PINK1 activation.
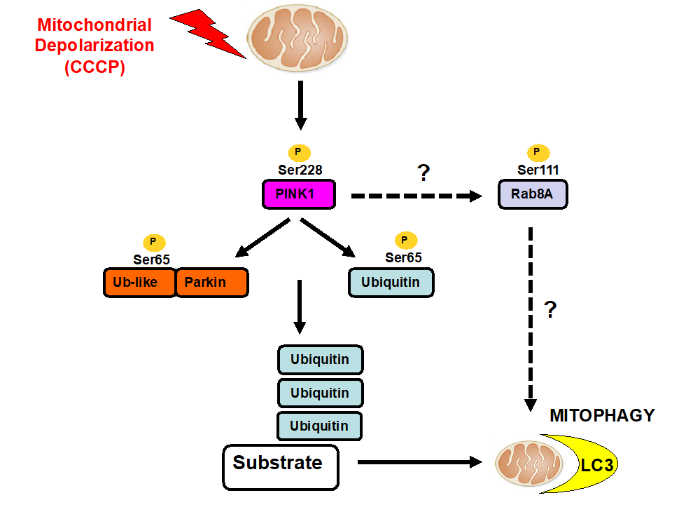
PINK1 kinase signalling networks in Parkinson's disease



