Overview
Decoding the Hidden Language of Ubiquitylation
Principal Investigator: Dr. Virginia De Cesare
Ubiquitylation is one of the cell’s most sophisticated regulatory mechanism - a molecular language that fine-tunes nearly every aspect of life, from protein stability to gene expression and stress responses. When this delicate mechanism is disrupted, the consequences reverberate across biology, inducing diseases such as cancer, neurodegeneration, and immunological disorders.
The attachment of ubiquitin to substrates is carried out by a cascade of enzymes: a single activating enzyme (E1) primes ubiquitin for transfer, before conjugation and ligation steps mediated by ubiquitin-conjugating enzymes (E2s) and E3 ligases (E3s) (Fig. 1). The human genome encodes ~40 E2s and more than 600 E3s, whose combinatorial and cooperative actions provide the specificity required to regulate thousands of substrates. RING E3 ligases represent the vast majority of known E3s, and they represent essential activators that facilitate the direct transfer of ubiquitin from the E2s to the substrate. E2s plays a fundamental role as they possess key activity determinants that direct the transfer of ubiquitin to the substrate and govern both the type of ubiquitin linkage and the extent of ubiquitin modification.
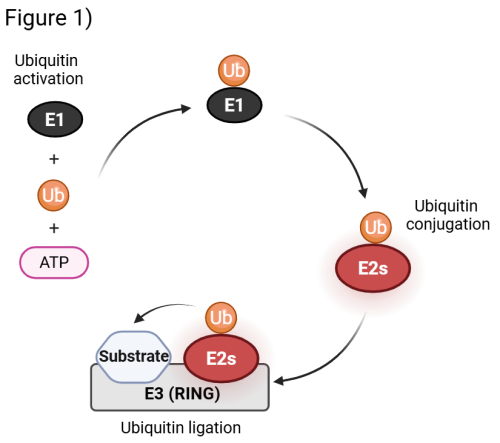
Exploring the Biology of Non-Canonical ubiquitylation
In the De Cesare lab, we investigate a paradigm-shifting dimension of ubiquitylation: non-canonical ubiquitylation. For decades, ubiquitin was thought to act mostly on protein substrates, attaching primarily to lysine residues. It is now clear, however, that this represents only part of the picture. Ubiquitin can also modify serine and threonine residues, as well as non-proteinaceous substrates such as nucleic acids (DNA and RNA) and even sugars. Unlike the stable isopeptide bond that mediates canonical ubiquitylation, non-canonical ubiquitylation is formed via oxyester bond, which is more labile and technically challenging to detect. This limited detectability has kept this field largely unexplored. As a result, the proteomic extent and biological role(s) of non-canonical ubiquitylation remain wide open for discovery - making it one of the most exciting frontiers in the ubiquitin and cell biology field (Fig. 2)
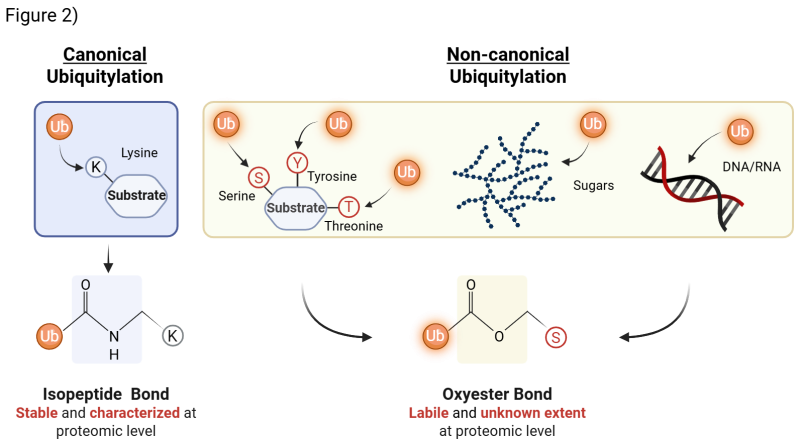
The De Cesare Lab has identified the UBE2Q family as a new class of non-canonical E2s capable of modifying substrates on serine and threonine residues - a foundational discovery that redefines how ubiquitylation extends beyond lysine modification [1]. This breakthrough is particularly significant because UBE2Q enzymes play crucial roles in fundamental biological processes, including embryo implantation, lysophagy, cancer progression, and neuronal survival, highlighting their broad impact on cellular regulation and human health.
Our Research Aims
Our research spans molecular, cellular, and translational levels, focusing on three interconnected goals (Fig. 3):
- Map the proteomic landscape of non-canonical ubiquitylation, revealing its interplay with other post-translational modifications and uncovering new layers of cellular regulation.
- Define the biological functions of non-canonical E2s, particularly UBE2Qs, identifying the pathways they regulate and their relevance to human health and disease, such as cancer and embryo implantation.
- Create state-of-the-art methods and biological tools to modulate non-canonical ubiquitylation, paving the way for novel therapeutic target identification and drug discovery.
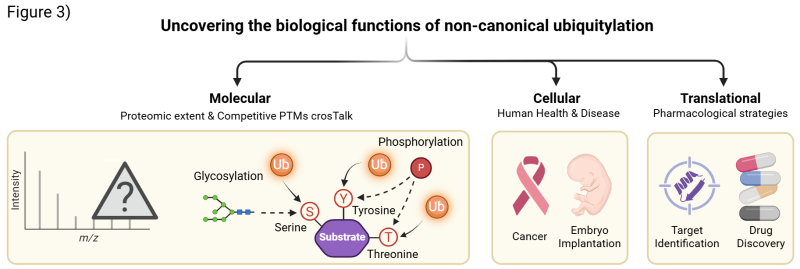
The lab investigates the biological functions of non-canonical ubiquitylation across molecular, cellular, and translational levels - from proteomic mapping and PTM cross-talk to disease mechanisms and therapeutic target discovery.
Our Approach and Environment
To tackle these questions, we integrate quantitative proteomics, molecular and cell biology, structural biology, and protein biochemistry. We also leverage a world-leading high-throughput MALDI-TOF/MS platform, which allows precise and scalable interrogation of ubiquitin enzymes [2, 3].
Our research sits at the intersection of discovery science and biomedical application, addressing fundamental questions that promise to reshape how we understand cellular regulation and disease.
Embedded within the MRC Protein Phosphorylation and Ubiquitylation Unit, our lab benefits from an exceptional collaborative research environment that bridges academia and industry.
How to join us
We are always seeking curious and motivated scientists at all career stages. If you are passionate about discovery and interested in contributing to a rapidly growing and cutting-edge area of research, we encourage you to contact the group leader at vdecesare@dundee.ac.uk to explore potential opportunities within the team.
References
1. Abdul Rehman, S.A., et al., Discovery and characterization of noncanonical E2-conjugating enzymes. Sci Adv, 2024. 10(13): p. eadh0123.
2. De Cesare, V., et al., Deubiquitinating enzyme amino acid profiling reveals a class of ubiquitin esterases. Proc Natl Acad Sci U S A, 2021. 118(4).
3. De Cesare, V., et al., High-throughput matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry-based deubiquitylating enzyme assay for drug discovery. Nat Protoc, 2020. 15(12): p. 4034-4057.



