
In a groundbreaking study, researchers from the MRC PPU, University of Dundee working in collaboration with researchers at University of Leeds have unveiled a critical new understanding of how cells control membrane protein synthesis. Published recently in Nature, this study illuminates a unique biochemical process known as UFMylation, revealing its essential role in maintaining cellular health.
Membrane-embedded and secreted proteins constitute about a third of all proteins synthesized in a cell. These proteins are made at the endoplasmic reticulum (ER) by ribosomes, the molecular machines that translate the genetic code into proteins. However, ribosomes can stall during this process, leading to collisions thereby disrupting normal protein synthesis. Cells employ quality control systems to recycle these ribosomes and eliminate the partially synthesized proteins. Failure to do so can lead to neurodegeneration. Quality control systems face challenges at the ER membrane, where ribosomes are associated with the translocon, the channel through which proteins are inserted into the membrane, making it difficult to access the ribosomes and the partially synthesized proteins. The mechanism by which ribosomes are separated from the translocon has been a long-standing mystery.
Central to this discovery is the process of UFMylation, in which a small ubiquitin-like protein, UFM1, is attached to the 60S ribosomal subunit protein, RPL26. This modification is a focus of the Kulathu lab. They recently showed that ribosome UFMylation is mediated by a highly unusual multiprotein ligase complex called the UFM1 Ribosome E3 Ligase (UREL) complex. To gain insights into this enigmatic process, Joshua Peter and Helge Magnussen developed a way to trap and purify the UREL complex in the act of attaching UFM1 to ribosomes. They collaborated with Elton Zeqiraj, a former PhD student at the MRC PPU and now a Professor at the University of Leeds, to image these complexes by cryo-electron microscopy. Linda Makhlouf at the Zeqiraj lab visualized the UREL complex bound to ribosomes in unprecedented detail.
The structure revealed UREL to form a C-shaped clamp around the 60S ribosomal subunit, blocking key sites essential for protein synthesis. Intriguingly, the findings suggest that UREL acts as a ‘writer’ to add UFM1 to ribosomes and then switches functions to become a ‘reader’ of the UFM1 modification it attached. As a ‘reader’ UREL inserts between the ribosome-translocon junction to wedge the ribosome free. Rohan Thakur, a PhD student in the lab, demonstrated that cells lacking UFMylation accumulate 60S-translocon complexes at the ER membrane. Together, these findings underscore the significant role for UFMylation in releasing stalled ribosomes from the endoplasmic reticulum (ER) membrane, facilitating the action of other quality control machinery to dispose of partially formed proteins and recycle ribosomes.
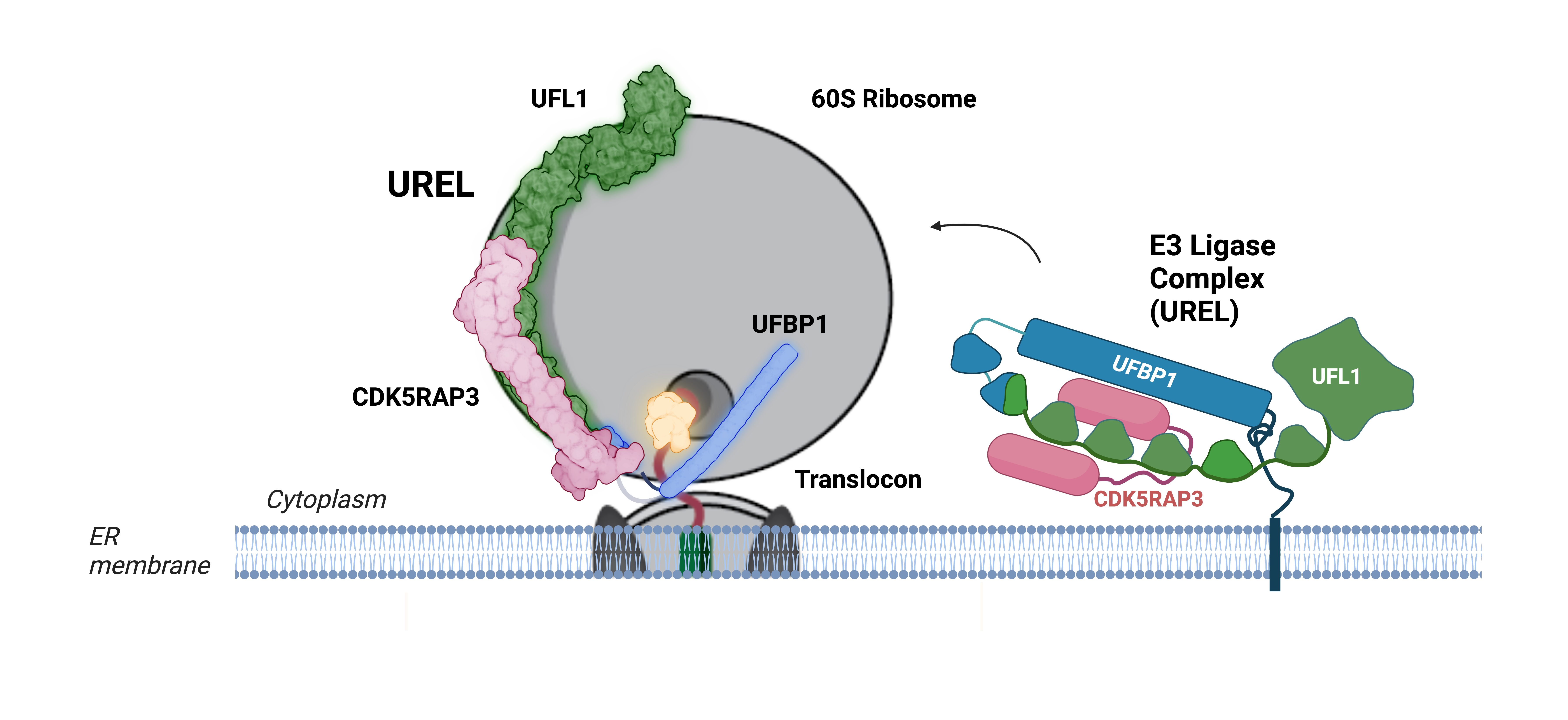
Schematic cartoon representation illustrating how the UFM1 ligase complex binds to 60S ribosomal subunit. (Figure was generated in Biorender) The ligase complex encircles the ribosome, forming a C-clamp-like architecture. One end of the complex (UFBP1, highlighted in blue) inserts itself into the junction between the ribosome and the translocon, wedging the 60S ribosomal subunit away from the translocon.
Despite being evolutionarily conserved, the function of UFMylation, has long remained enigmatic. This research from the Kulathu lab elucidates a fundamental function of UFMylation. It also reveals for the first time the unique characteristics of the UFM1 E3 ligase complex. Helge Magnussen, a postdoctoral researcher in the lab, determined crystal structures of sub-complexes. Together with cross-linking mass spectrometry performed in collaboration with Anton Calabrese’s lab, these structures revealed that the ligase employs a multistep process to attach UFM1 to its substrates.
This work clarifies why the UFM1 system is present in most eukaryotes and underscores the importance of UFMylation in maintaining cellular health. The association of muations in the UFM1 system with a wide range of pathologies, including cerebellar ataxia, neurodevelopmental disorders, and skeletal abnormalities, underlines the significance of UFMylation and the potential for novel therapeutic strategies. This discovery represents a substantial leap forward in our understanding of cellular biology. It illuminates the intricate mechanisms cells use to maintain protein homeostasis and paves the way for further research into the quality control pathways orchestrated by UFMylation at the ER membrane.
This work was funded by the UKRI MRC, BBSRC, the Wellcome Trust, the Lister Institute for Preventive Medicine and the European Research Council (ERC).
See more:
The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons Mahklouf L, Peter JJ, Magnussen H, Thakur R, Millrine D, Minshull TC, Harrison G, Varghese J, Lamoliatte F, Foglizzo M, Macartney T, Calabrese AN, Zeqiraj E and Kulathu Y. Nature
UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER DaRosa PA, Penchev I, Gumbin SC, Scavone F, Wąchalska M, Paulo JA, Ordureau A, Peter JJ, Kulathu Y, Harper JW, Becker T, Beckmann R and Kopito RR. Nature

