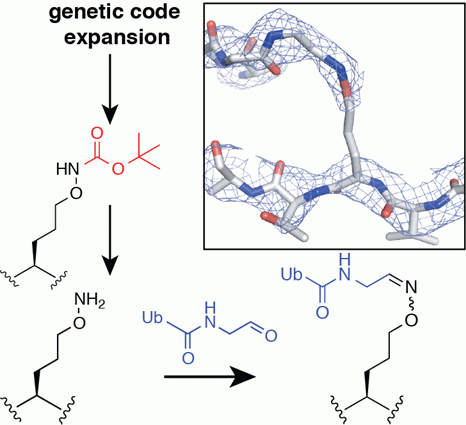
Researchers in the laboratory of Dr. Satpal Virdee have published the first example of genetically encodable aminooxy functionality, which can be installed site-selectively into recombinant proteins using genetic code expansion technology. The findings were published in Chembiochem this week.
Site-specific modification of proteins is a challenging and hotly pursued topic within the chemical biology community. The ability to direct site-selective chemical bond formation between proteins in the presence of chemically diverse functionality within biological samples is non-trivial but advantages and insight gained from such methodologies are indispensible to modern biology. Current focus is on developing ever faster chemistries whilst retaining biorthogonality.
One example of a bioorthogonal reaction is the oxime ligation reaction between an aldehyde (or ketone) with an aminooxy (-ONH2) functional group. However, methods that allow installation of the prerequisite chemical handles for oxime ligation at the genetic level into proteins are limited.
The installation of this aminooxy functional group into ubiquitin, and into the ubiquitin-like protein SUMO2, enabled researchers to carry out site-specific 'chemical ubiquitylation' of full-length proteins to produce diubiquitin conjugates and a ubiquitylated substrate (ubiquitylated SUMO2) as proof-of-principle. Importantly, the oxime linkage produced between the protein molecules as a result of the ligation reaction is hydrolytically stable to deubiquitinase (DUB) proteolytic activity whilst being an exquisite structural mimic of the native isopeptide bond which it successfully substitutes.
The technology has enabled Virdee lab researchers to generate a valuable protein toolkit, including protein-based nanomolar DUB inhibitors and non-hydrolysable extended ubiquitin polymers which now provide researchers with a means to probe the cellular roles of Ub linkages without the intrinsic sensitivity of native ubiquitin linkages to DUB activity and to potentially identify hitherto unknown proteins that interact with ubiquitylated substrates and distinct ubiquitin chain types.
The first author of the study, Mathew Stanley, describes the potential use of these tools developed in the Virdee lab. 'The protein conjugates produced using this method are structurally highly similar to native conjugates as determined by biophysical techniques and X-ray crystallography. Coupled with the fact that they are stable to the hydrolytic activity of DUBs make these invaluable tools for studying ubiquitin linkage specific processes in cells and for the study of DUB biology, which has become a significant area of interest for many pharmaceutical companies. Looking beyond the ubiquitin system, because of the modularity and versatility of the approach, the methods described will have wide ranging utility in other areas, for example, the biochemical and structural study of other families of proteolytic enzymes and in the production of antibody-drug conjugates and biomaterials.'
A light activatable variant of the aminooxy functional group that the research team present have the group particularly excited about the future applications of the technology.
'With recent advances in ultra-fast oxime labeling reactions and with the existing ability to genetically encode aldehyde functionality into proteins, our contribution now paves the way towards a strategy for light-dependent ultrafast protein labelling and site-specific protein–protein tethering in live cells'.

